-
Notifications
You must be signed in to change notification settings - Fork 2.7k
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
[Project] Medical semantic seg dataset: bccs (#2861)
- Loading branch information
1 parent
b8b6ee6
commit 4feba31
Showing
7 changed files
with
285 additions
and
0 deletions.
There are no files selected for viewing
123 changes: 123 additions & 0 deletions
123
projects/medical/2d_image/histopathology/breastCancerCellSegmentation/README.md
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,123 @@ | ||
| # breastCancerCellSegmentation | ||
|
|
||
| ## Description | ||
|
|
||
| This project supports **`breastCancerCellSegmentation`**, which can be downloaded from [here](https://www.heywhale.com/mw/dataset/5e9e9b35ebb37f002c625423). | ||
|
|
||
| ### Dataset Overview | ||
|
|
||
| This dataset, with 58 H&E-stained histopathology images was used for breast cancer cell detection and associated real-world data. | ||
| Conventional histology uses a combination of hematoxylin and eosin stains, commonly referred to as H&E. These images are stained because most cells are inherently transparent with little or no intrinsic pigment. | ||
| Certain special stains selectively bind to specific components and can be used to identify biological structures such as cells. | ||
|
|
||
| ### Original Statistic Information | ||
|
|
||
| | Dataset name | Anatomical region | Task type | Modality | Num. Classes | Train/Val/Test Images | Train/Val/Test Labeled | Release Date | License | | ||
| | -------------------------------------------------------------------------------------------- | ----------------- | ------------ | -------------- | ------------ | --------------------- | ---------------------- | ------------ | --------------------------------------------------------------- | | ||
| | [breastCancerCellSegmentation](https://www.heywhale.com/mw/dataset/5e9e9b35ebb37f002c625423) | cell | segmentation | histopathology | 2 | 58/-/- | yes/-/- | 2020 | [CC-BY-NC 4.0](https://creativecommons.org/licenses/by-sa/4.0/) | | ||
|
|
||
| | Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test | | ||
| | :--------------: | :--------: | :--------: | :------: | :------: | :-------: | :-------: | | ||
| | background | 58 | 98.37 | - | - | - | - | | ||
| | breastCancerCell | 58 | 1.63 | - | - | - | - | | ||
|
|
||
| Note: | ||
|
|
||
| - `Pct` means percentage of pixels in this category in all pixels. | ||
|
|
||
| ### Visualization | ||
|
|
||
| 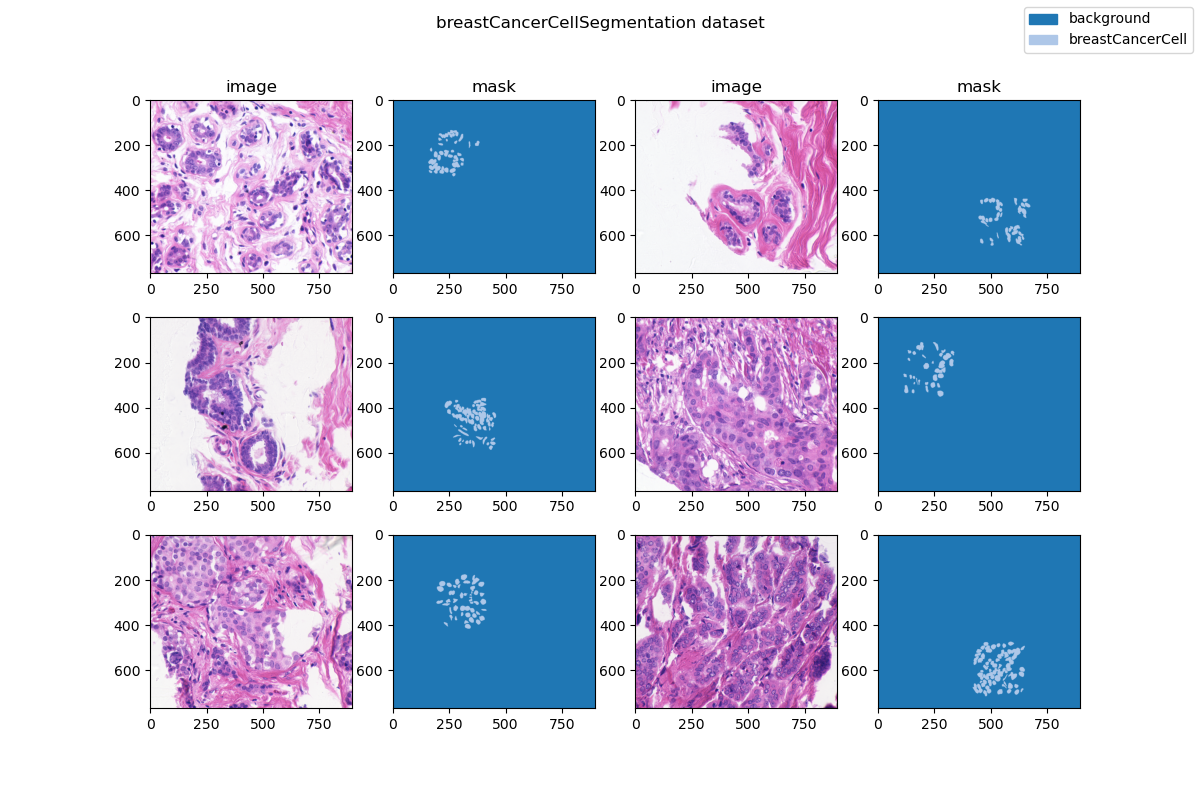 | ||
|
|
||
| ## Usage | ||
|
|
||
| ### Prerequisites | ||
|
|
||
| - Python v3.8 | ||
| - PyTorch v1.10.0 | ||
| - pillow (PIL) v9.3.0 | ||
| - scikit-learn (sklearn) v1.2.0 | ||
| - [MIM](https://github.com/open-mmlab/mim) v0.3.4 | ||
| - [MMCV](https://github.com/open-mmlab/mmcv) v2.0.0rc4 | ||
| - [MMEngine](https://github.com/open-mmlab/mmengine) v0.2.0 or higher | ||
| - [MMSegmentation](https://github.com/open-mmlab/mmsegmentation) v1.0.0 | ||
|
|
||
| All the commands below rely on the correct configuration of `PYTHONPATH`, which should point to the project's directory so that Python can locate the module files. In `breastCancerCellSegmentation/` root directory, run the following line to add the current directory to `PYTHONPATH`: | ||
|
|
||
| ```shell | ||
| export PYTHONPATH=`pwd`:$PYTHONPATH | ||
| ``` | ||
|
|
||
| ### Dataset Preparing | ||
|
|
||
| - Download dataset from [here](https://www.heywhale.com/mw/dataset/5e9e9b35ebb37f002c625423) and save it to the `data/` directory . | ||
| - Decompress data to path `data/`. This will create a new folder named `data/breastCancerCellSegmentation/`, which contains the original image data. | ||
| - run script `python tools/prepare_dataset.py` to format data and change folder structure as below. | ||
|
|
||
| ```none | ||
| mmsegmentation | ||
| ├── mmseg | ||
| ├── projects | ||
| │ ├── medical | ||
| │ │ ├── 2d_image | ||
| │ │ │ ├── histopathology | ||
| │ │ │ │ ├── breastCancerCellSegmentation | ||
| │ │ │ │ │ ├── configs | ||
| │ │ │ │ │ ├── datasets | ||
| │ │ │ │ │ ├── tools | ||
| │ │ │ │ │ ├── data | ||
| │ │ │ │ │ │ ├── breastCancerCellSegmentation | ||
| | │ │ │ │ │ │ ├── train.txt | ||
| | │ │ │ │ │ │ ├── val.txt | ||
| | │ │ │ │ │ │ ├── images | ||
| | │ │ │ │ │ │ | ├── xxx.tif | ||
| | │ │ │ │ │ │ ├── masks | ||
| | │ │ │ │ │ │ | ├── xxx.TIF | ||
| ``` | ||
|
|
||
| ### Training commands | ||
|
|
||
| Train models on a single server with one GPU. | ||
|
|
||
| ```shell | ||
| mim train mmseg ./configs/${CONFIG_FILE} | ||
| ``` | ||
|
|
||
| ### Testing commands | ||
|
|
||
| Test models on a single server with one GPU. | ||
|
|
||
| ```shell | ||
| mim test mmseg ./configs/${CONFIG_FILE} --checkpoint ${CHECKPOINT_PATH} | ||
| ``` | ||
|
|
||
| ## Checklist | ||
|
|
||
| - [x] Milestone 1: PR-ready, and acceptable to be one of the `projects/`. | ||
|
|
||
| - [x] Finish the code | ||
|
|
||
| - [x] Basic docstrings & proper citation | ||
|
|
||
| - [x] Test-time correctness | ||
|
|
||
| - [x] A full README | ||
|
|
||
| - [ ] Milestone 2: Indicates a successful model implementation. | ||
|
|
||
| - [ ] Training-time correctness | ||
|
|
||
| - [ ] Milestone 3: Good to be a part of our core package! | ||
|
|
||
| - [ ] Type hints and docstrings | ||
|
|
||
| - [ ] Unit tests | ||
|
|
||
| - [ ] Code polishing | ||
|
|
||
| - [ ] Metafile.yml | ||
|
|
||
| - [ ] Move your modules into the core package following the codebase's file hierarchy structure. | ||
|
|
||
| - [ ] Refactor your modules into the core package following the codebase's file hierarchy structure. |
42 changes: 42 additions & 0 deletions
42
...stopathology/breastCancerCellSegmentation/configs/breastCancerCellSegmentation_512x512.py
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,42 @@ | ||
| dataset_type = 'breastCancerCellSegmentationDataset' | ||
| data_root = 'data/breastCancerCellSegmentation' | ||
| img_scale = (512, 512) | ||
| train_pipeline = [ | ||
| dict(type='LoadImageFromFile', imdecode_backend='tifffile'), | ||
| dict(type='LoadAnnotations', imdecode_backend='tifffile'), | ||
| dict(type='Resize', scale=img_scale, keep_ratio=False), | ||
| dict(type='RandomFlip', prob=0.5), | ||
| dict(type='PhotoMetricDistortion'), | ||
| dict(type='PackSegInputs') | ||
| ] | ||
| test_pipeline = [ | ||
| dict(type='LoadImageFromFile', imdecode_backend='tifffile'), | ||
| dict(type='Resize', scale=img_scale, keep_ratio=False), | ||
| dict(type='LoadAnnotations', imdecode_backend='tifffile'), | ||
| dict(type='PackSegInputs') | ||
| ] | ||
| train_dataloader = dict( | ||
| batch_size=16, | ||
| num_workers=4, | ||
| persistent_workers=True, | ||
| sampler=dict(type='InfiniteSampler', shuffle=True), | ||
| dataset=dict( | ||
| type=dataset_type, | ||
| data_root=data_root, | ||
| ann_file='train.txt', | ||
| data_prefix=dict(img_path='images', seg_map_path='masks'), | ||
| pipeline=train_pipeline)) | ||
| val_dataloader = dict( | ||
| batch_size=1, | ||
| num_workers=4, | ||
| persistent_workers=True, | ||
| sampler=dict(type='DefaultSampler', shuffle=False), | ||
| dataset=dict( | ||
| type=dataset_type, | ||
| data_root=data_root, | ||
| ann_file='val.txt', | ||
| data_prefix=dict(img_path='images', seg_map_path='masks'), | ||
| pipeline=test_pipeline)) | ||
| test_dataloader = val_dataloader | ||
| val_evaluator = dict(type='IoUMetric', iou_metrics=['mIoU', 'mDice']) | ||
| test_evaluator = dict(type='IoUMetric', iou_metrics=['mIoU', 'mDice']) |
18 changes: 18 additions & 0 deletions
18
...ion/configs/fcn-unet-s5-d16_unet_1xb16-0.0001-20k_breastCancerCellSegmentation-512x512.py
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,18 @@ | ||
| _base_ = [ | ||
| 'mmseg::_base_/models/fcn_unet_s5-d16.py', | ||
| './breastCancerCellSegmentation_512x512.py', | ||
| 'mmseg::_base_/default_runtime.py', | ||
| 'mmseg::_base_/schedules/schedule_20k.py' | ||
| ] | ||
| custom_imports = dict(imports='datasets.breastCancerCellSegmentation_dataset') | ||
| img_scale = (512, 512) | ||
| data_preprocessor = dict(size=img_scale) | ||
| optimizer = dict(lr=0.0001) | ||
| optim_wrapper = dict(optimizer=optimizer) | ||
| model = dict( | ||
| data_preprocessor=data_preprocessor, | ||
| decode_head=dict(num_classes=2), | ||
| auxiliary_head=None, | ||
| test_cfg=dict(mode='whole', _delete_=True)) | ||
| vis_backends = None | ||
| visualizer = dict(vis_backends=vis_backends) |
18 changes: 18 additions & 0 deletions
18
...tion/configs/fcn-unet-s5-d16_unet_1xb16-0.001-20k_breastCancerCellSegmentation-512x512.py
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,18 @@ | ||
| _base_ = [ | ||
| 'mmseg::_base_/models/fcn_unet_s5-d16.py', | ||
| './breastCancerCellSegmentation_512x512.py', | ||
| 'mmseg::_base_/default_runtime.py', | ||
| 'mmseg::_base_/schedules/schedule_20k.py' | ||
| ] | ||
| custom_imports = dict(imports='datasets.breastCancerCellSegmentation_dataset') | ||
| img_scale = (512, 512) | ||
| data_preprocessor = dict(size=img_scale) | ||
| optimizer = dict(lr=0.001) | ||
| optim_wrapper = dict(optimizer=optimizer) | ||
| model = dict( | ||
| data_preprocessor=data_preprocessor, | ||
| decode_head=dict(num_classes=2), | ||
| auxiliary_head=None, | ||
| test_cfg=dict(mode='whole', _delete_=True)) | ||
| vis_backends = None | ||
| visualizer = dict(vis_backends=vis_backends) |
18 changes: 18 additions & 0 deletions
18
...ation/configs/fcn-unet-s5-d16_unet_1xb16-0.01-20k_breastCancerCellSegmentation-512x512.py
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,18 @@ | ||
| _base_ = [ | ||
| 'mmseg::_base_/models/fcn_unet_s5-d16.py', | ||
| './breastCancerCellSegmentation_512x512.py', | ||
| 'mmseg::_base_/default_runtime.py', | ||
| 'mmseg::_base_/schedules/schedule_20k.py' | ||
| ] | ||
| custom_imports = dict(imports='datasets.breastCancerCellSegmentation_dataset') | ||
| img_scale = (512, 512) | ||
| data_preprocessor = dict(size=img_scale) | ||
| optimizer = dict(lr=0.01) | ||
| optim_wrapper = dict(optimizer=optimizer) | ||
| model = dict( | ||
| data_preprocessor=data_preprocessor, | ||
| decode_head=dict(num_classes=2), | ||
| auxiliary_head=None, | ||
| test_cfg=dict(mode='whole', _delete_=True)) | ||
| vis_backends = None | ||
| visualizer = dict(vis_backends=vis_backends) |
30 changes: 30 additions & 0 deletions
30
...topathology/breastCancerCellSegmentation/datasets/breastCancerCellSegmentation_dataset.py
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,30 @@ | ||
| from mmseg.datasets import BaseSegDataset | ||
| from mmseg.registry import DATASETS | ||
|
|
||
|
|
||
| @DATASETS.register_module() | ||
| class breastCancerCellSegmentationDataset(BaseSegDataset): | ||
| """breastCancerCellSegmentationDataset dataset. | ||
| In segmentation map annotation for breastCancerCellSegmentationDataset, | ||
| ``reduce_zero_label`` is fixed to False. The ``img_suffix`` | ||
| is fixed to '.png' and ``seg_map_suffix`` is fixed to '.png'. | ||
| Args: | ||
| img_suffix (str): Suffix of images. Default: '.png' | ||
| seg_map_suffix (str): Suffix of segmentation maps. Default: '.png' | ||
| reduce_zero_label (bool): Whether to mark label zero as ignored. | ||
| Default to False. | ||
| """ | ||
| METAINFO = dict(classes=('background', 'breastCancerCell')) | ||
|
|
||
| def __init__(self, | ||
| img_suffix='_ccd.tif', | ||
| seg_map_suffix='.TIF', | ||
| reduce_zero_label=False, | ||
| **kwargs) -> None: | ||
| super().__init__( | ||
| img_suffix=img_suffix, | ||
| seg_map_suffix=seg_map_suffix, | ||
| reduce_zero_label=reduce_zero_label, | ||
| **kwargs) |
36 changes: 36 additions & 0 deletions
36
...cts/medical/2d_image/histopathology/breastCancerCellSegmentation/tools/prepare_dataset.py
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,36 @@ | ||
| import argparse | ||
| import glob | ||
| import os | ||
|
|
||
| from sklearn.model_selection import train_test_split | ||
|
|
||
|
|
||
| def save_anno(img_list, file_path, suffix): | ||
| # 只保留文件名,不保留后缀 | ||
| img_list = [x.split('/')[-1][:-len(suffix)] for x in img_list] | ||
|
|
||
| with open(file_path, 'w') as file_: | ||
| for x in list(img_list): | ||
| file_.write(x + '\n') | ||
|
|
||
|
|
||
| if __name__ == '__main__': | ||
| parser = argparse.ArgumentParser() | ||
| parser.add_argument( | ||
| '--data_root', default='data/breastCancerCellSegmentation/') | ||
| args = parser.parse_args() | ||
| data_root = args.data_root | ||
|
|
||
| # 1. 划分训练集、验证集 | ||
| # 1.1 获取所有图片路径 | ||
| img_list = glob.glob(os.path.join(data_root, 'images', '*.tif')) | ||
| img_list.sort() | ||
| mask_list = glob.glob(os.path.join(data_root, 'masks', '*.TIF')) | ||
| mask_list.sort() | ||
| assert len(img_list) == len(mask_list) | ||
| # 1.2 划分训练集、验证集、测试集 | ||
| train_img_list, val_img_list, train_mask_list, val_mask_list = train_test_split( # noqa | ||
| img_list, mask_list, test_size=0.2, random_state=42) | ||
| # 1.3 保存划分结果 | ||
| save_anno(train_img_list, os.path.join(data_root, 'train.txt'), '_ccd.tif') | ||
| save_anno(val_img_list, os.path.join(data_root, 'val.txt'), '_ccd.tif') |